The study of cytokines is valuable in investigating inflammation. Cytokines are small proteins that transmit signals from cell to cell and trigger immunity to combat infection, inflammation and other disorders. Accurate detection and quantification of cytokine concentrations, therefore, becomes essential in describing the nature of the infection and powerful to follow the disease-staging process.
Enzyme labels and immunoassays using enzyme-conjugated antibodies have become increasingly popular among scientists. The indirect sandwich enzyme-linked immunosorbent assay or ELISA is a cost-effective analytical tool commonly used for antibodies, antigens, proteins and glycoproteins analysis. Applications for cytokine analysis using ELISA are widespread, including those for neurological diseases.
Our experts at Helvetica Health Care (HHC) highlight this powerful medical application and how it helps in cytokine analysis.
What is ELISA, and why is it popular for cytokine detection?
The traditional Sandwich ELISA test is a simple yet widely used tool in medical laboratories worldwide for detecting, quantifying, and discriminating a single cytokine from several biomolecules in any given sample.
ELISAs produce specific, precise and sensitive results, with high-throughput rates necessary for cytokine protein analysis whether the study is in vitro cell culture system or an in vivo animal model. Typically, ELISA can measure cytokines produced in a culture medium, the concentration of ex vivo cytokine levels in plasma and serum, and induced cytokine production in our whole blood assays.
ELISA provides high accuracy because it uses antibodies specific to one cytokine or form. Sequential ELISA assays are used to analyse multiple cytokines proteins from samples with small volumes.
How do ELISAs work?
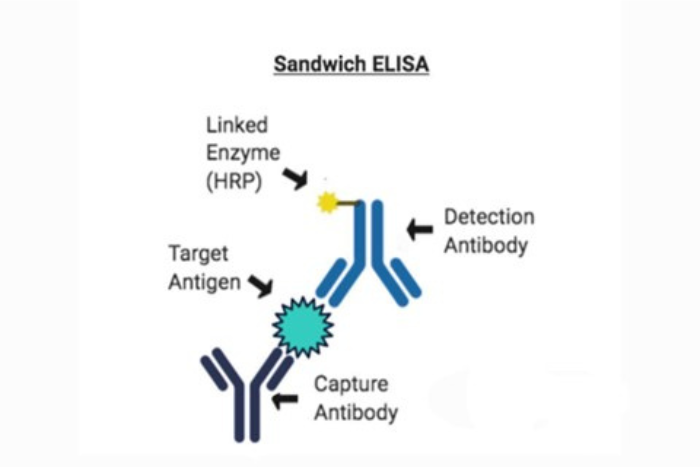
In principle, a basic sandwich ELISA analyses antigens between a layer of capture and detection antibodies. In other words, it is performed using an antibody-antigen-antibody sandwich. Below we explain the four steps of the simple indirect sandwich ELISA process:
1. Extracting analyte (specimen extracted for analysis) from a capture antibody sample.
2. Identifying biotin-labelled captured analyte with detection antibody (also specific for extracted analyte).
3. Detection amplification with streptavidin conjugated with an enzyme
4. Adding substrate and measuring the coloured signal via optical density (OD) using a microplate reader.
Scientists frequently use a microwell format (microwell plates) when conducting a traditional ELISA test.
Before we move on to a detailed explanation of the above steps, let us remind you that HHC provides a wide range of ELISA kits ready to use with break-apart wells. Our range of ELISA includes:
• The RETROTEK™ range is designed for the detection and quantitation of retroviral antigens from the retroviruses HIV-1, SIV and HTLV found in cell culture, serum, plasma or other biological fluids
• IMMUNOTEK™ kits can detect and quantify various immunoglobulins from many species, including Humans, Chicken, Cow, Goat, Rabbit, Rat and Mouse. We also provide HHV-6 IgG antibody and KSHV/HHV8 IgG antibody ELISA kits.
The General ELISA method explained
Typically, the sample contains anti-cytokine antibodies that are highly purified, also known as capture antibodies, linked to an enzyme. Post plate washings, the immobilised antibodies are used to capture the soluble cytokine proteins present in the applied samples. After washing unbound material, biotin-conjugated anti-cytokine antibodies (detection antibodies) are used to identify the captured cytokine proteins.
Next is an enzyme-labelled avidin or streptavidin stage. When a chromogenic substrate is added to the capture antibodies, they produce a signal detected due to the substrate’s colour change. The amount of coloured product generated by the bound, enzyme-linked detection reagents is easily detected spectrophotometrically using an ELISA-plate reader at the proper optical density (OD). Data storage and reanalysis are significantly facilitated when the plate reader is connected to a computer.
By serially diluting a standard cytokine protein solution with a defined concentration, a standard curve is added to a sandwich ELISA experiment. Standard curves, often referred to as calibration curves, are typically plotted as the standard cytokine protein concentration (commonly ng or pg of cytokine/ml) vs the associated mean OD value of duplicates. It is possible to interpolate the concentrations of the putative cytokine-containing samples from the standard curve.
The use of an ELISA computer application simplifies this operation. In general, performing a dilution series of the unknown samples is helpful to ensure that the OD will fall within the linear range of the standard curve. The type of ELISA reagents used may influence the curve fit analysis that researchers utilise on their data, including either linear-log, log-log, or four-parameter transformations.
Just like there are different types of cytokines, there exist other ELISAs procedures too:
- The Checkerboard method for optimising antibody concentrations
- The Dose-Response method for optimal blocking and dilution buffer for the sample matrix
- The Standard ELISA method for determining an analyte’s concentration in a given sample
- The Sequential ELISA
- The Multiplex ELISA
This basic technique can be enhanced using the sequential ELISA to assess multiple analytes from a single sample aliquot. In this case, a single sample is extracted from one ELISA plate and then incubated in a separate plate as the first plate would only capture the cytokine detected by the antibody utilised for capturing the cytokine.
Multiplex ELISA is an excellent alternate method to the sequential ELISA wherein numerous capture antibodies are printed into a single microplate well, each with a different set of specificities.
To know the availability of our ELISA kits, contact us at our office in Geneva.